What in the World is Going on with Forensic DNA Mixture Analysis?

Suzanna Ryan,
MS, D-ABC
Owner/Consultant at Ryan Forensic DNA Consulting.
If you are even remotely involved in the world of criminal investigations, whether it be as an investigator, attorney, or scientist, you have surely heard or read one or more articles about issues surrounding DNA mixture interpretation. However, many of the news stories I read don’t fully explain these issues, leaving many to try to guess what exactly is happening in forensic laboratories. Forensic laboratories are using only the best petri trays, microscopes and individually wrapped serological pipets to discover more about DNA. This article seeks to explain and answer the issues surrounding forensic DNA mixture interpretation.
DNA Mixtures
For some time it has been recognized by many forensic experts that mixtures were a problem area in the DNA field. For example, a large inter-laboratory study conducted by Dr. John Butler and Dr. Margaret Klein of the National Institute of Standards and Technology (NIST) in 2005 found that even when trained DNA analysts are looking at the same data, different conclusions and different statistical results were reached. Even when all measures are taken to ensure uncontaminated specimen during tests, such as ventilation controlled workspaces and use of biosafety equipment like a biological cabinet with intuitive user-friendly control panel that usually act to prevent contamination and provide precise results, there was significant difference in reported conclusions. The study found that laboratories showed a difference of 10 orders of magnitude in their statistical conclusions (ranging from 105 to 1015) based on which alleles were deduced and reported. The state of DNA mixture interpretations in forensic laboratories led the esteemed Peter Gill, formerly of the UK’s Forensic Science Service to opine that, “If you show 10 colleagues a mixture, you will probably end up with 10 different answers”.
Many outside the forensic community first heard about mixture issues in August 2015 when the Texas Forensic Science Commission issued a statement detailing the problems that were uncovered in various Texas laboratories after the FBI issued its amendments to their DNA population databases. Many people conflated the two issues, however, the FBI database errors and DNA mixture interpretation are actually two completely separate issues. When the FBI released their amended database, labs in Texas began re-doing the statistical conclusions on prior cases at attorney’s request. Part of this process involved re-interpreting mixtures of DNA using new protocols and procedures that were put into place following the implementation of the 2010 Scientific Working Group on DNA Analysis Methods (SWGDAM) Interpretation Guidelines for Autosomal STR Typing by Forensic DNA Testing Laboratories. It was then that the real issue was discovered.
When some of the more complex mixtures were re-interpreted using the new guidelines, it was discovered that there could be vast differences in the statistical conclusions. In fact, it was now quite possible that a sample previously reported as an inclusion (meaning the person of interest was found to be included as a contributor to the mixture) might very well now be reported as inconclusive (meaning that the lab could no longer include the same person of interest as a contributor to the mixture). Of course these changes made the defense community in Texas sit up and take notice. Texas has been quite proactive with their re-analysis of older cases. This is certainly not the case in the vast majority of other states and jurisdictions.
What’s the Problem?
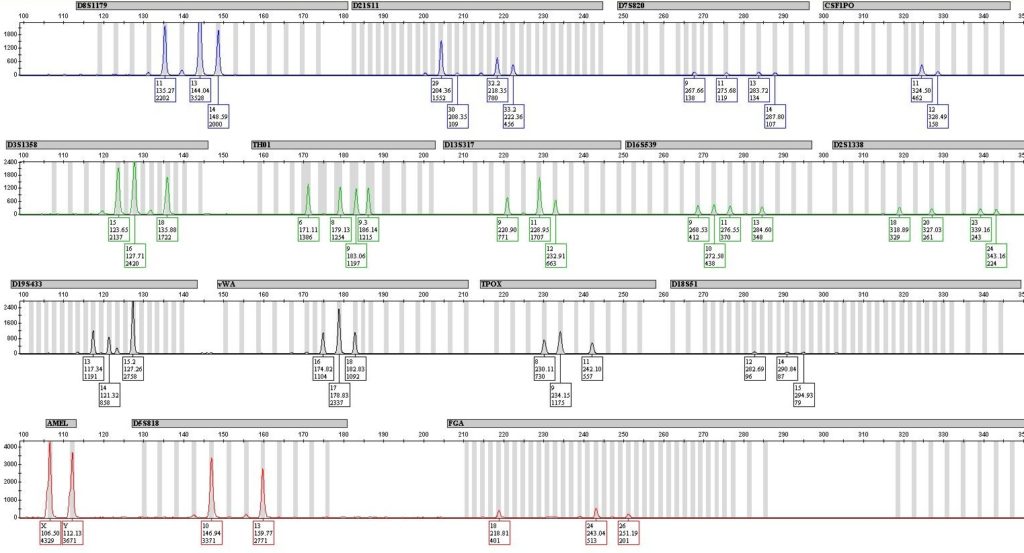
So what are the issues with mixture interpretation? The main issue revolves around the use of the Combined Probability of Inclusion (CPI) statistic. While the CPI calculation is not problematic in and of itself, it is the way many labs were applying this statistic to low-level data that became problematic. CPI is designed to give the user an answer to the question “given this set of DNA types at these DNA locations, what is the probability that another, unrelated, individual other than the person of interest, could also be a contributor to the mixture”. In other words, if I went out and randomly selected a set number of individuals, how many individuals would I need to test until I found another person who had DNA types in their profile that are present in the mixture found on the given item of evidence.
If all of the possible DNA types are present at a significant level, and there are no indications of additional DNA types below the lab’s reporting threshold, then there is no issue with the CPI statistic. But, many samples can yield complex mixture profiles originating from very minimal amounts of DNA.
If low level DNA types are present, this is a sign the sample may be suffering from stochastic effects – basically, random fluctuations that can occur during the copying step of the process – and this means that there could be missing DNA types. If not all of the data is present, then not all of the genotypes are represented – you can find out more about genotyping at LifeNet Health for example. If not all of the genotypes are represented, the CPI statistic is not valid. Dr. John Butler from NIST, author of numerous DNA textbooks, including what is generally referred to as the “DNA Bible” by lab analysts, has stated repeatedly in his talks, books, and other presentations that the CPI statistic cannot handle dropout and therefore should not be used in unrestricted CPI calculations.
In an effort to help ensure that only the loci where all alleles are present are actually used in the statistical calculations, SWGDAM, in 2010, recommended the use of two threshold, the analytical threshold and the stochastic threshold. The analytical threshold is the height that a possible DNA type much reach before the lab considers the peak to be a “true” DNA peak, and not just noise or some sort of amplification artifact. The stochastic threshold has been defined by SWGDAM as, “the peak height value above which it is reasonable to assume that, at a given locus, allelic dropout of a sister allele has not occurred”. This basically means that the lab must set a stochastic threshold, based upon the validation of the particular instrument and amplification kit in use, that provides them with a level of confidence such that they can be certain that drop-out of data (alleles) has not occurred due to low levels of input material.
SWGDAM’s Impact
The introduction of the stochastic threshold had a drastic impact on complex mixtures which were evaluated using the CPI statistic. Now, lab analysts were required to evaluate a mixture profile prior to comparison with the known reference samples and make a determination regarding which loci were useful for comparison purposes. If the lab did not have addition guidelines allowing them to group together “major clusters” (called a restricted CPI calculation), or to assume certain numbers of contributors thus allowing them to perform a different type of statistical calculation (termed a restricted Random Match Probability), then any locus which contained even a single allele that was below the lab’s stochastic threshold could not be used for statistical calculations. Any locus with low level data had to be disregarded when it came time to compare the results to reference samples. Often that meant that instead of including a person of interest at all or most of the loci, the analyst might now have to report that only 2 or 3 of the loci contained all alleles above the stochastic threshold and therefore only those two or three loci were useful for statistical purposes, hence the drastic changes in some cases of the CPI statistic.
There are other issues that were addressed by the 2010 SWGDAM guidelines, including SWGDAM’s guidance that the profile should be interpreted prior to a comparison with any known reference samples. The main reason for this guideline is to help reduce any potential unintentional bias from occurring during the interpretation of the mixed DNA profile. Researchers Itiel Dror and Greg Hampikian showed in their 2011 Science and Justice article that mixture interpretation could be quite subjective when contextual information – such as knowing that an individual was considered a suspect in the case and had been implicated by a co-defendant – was introduced. In their study, 17 qualified DNA analysts examined the same data (which had previously been analyzed in an adjudicated court case wherein the suspect was included as a possible contributor to the profile). Of these 17 examiners, only 1 reached the same conclusion as the original analyst. Twelve analysts excluded the suspect and 4 reached an “inconclusive”‘ conclusion.
“Suspect-Centric” Statistical Analysis
Prior to the SWGDAM changes with most unresolvable mixtures, lab analysts would make a direct comparison of known reference samples to the mixture profile developed from the evidence. This direct comparison often led to different CPI statistics being reported for different references on the same evidence sample. So, one suspect might have a CPI of 1 in 1,000 while a second suspect might have a CPI of 1 in 1,000,000 for the same item of evidence. This method has been determined to be a suspect-centric method that is not conservative and could, in fact, be overly-inclusive. The loci useful for comparison purposes, and the alleles that are part of any major or minor contributors, should be determined prior to any comparisons with known reference samples. In this way, a single CPI could be calculated using the eligible loci/alleles prior to any comparison with a reference sample. The reference sample then is either included in the possible combination of alleles detected at a particular locus, or it is excluded as a possible contributor. As stated by Bieber et al “adjustments to fit the interpretation to reject or reinstate a locus based on additional information from a person of interest’s profile (i.e. confirmation bias and fitting the profile interpretation to explain missing data based on a known sample) are inappropriate.”
Another common practice in regards to the CPI calculation was the practice of dropping those loci from the statistical calculation where the person of interest was not fully represented. This has often been cited as a “conservative” method of analysis. However, this method has been shown by statisticians James Curran and John Buckleton to be anything but conservative. They found that the risk of this method “producing apparently strong evidence against an innocent suspect was “not negligible”. As an example, say that there are two loci present in a mixture profile which do not contain the DNA types of the person of interest. Instead of eliminating the person of interest or determining that the profile was inconclusive, lab analysts would often simply “drop” the affected loci from the calculation in order to effectively “make” the data fit the person of interest’s profile.
The vast majority of labs changed their mixture interpretation guidelines soon after 2010. Those labs that did not appropriately change their guidelines, like the DC Consolidated Laboratory, the Broward County Sheriff’s Office, and the Austin Police Department laboratory, have gained unwanted national attention through their temporary or permanent closure and loss of accreditation.
What About All the Cases Interpreted Before the SWGDAM Changes?
However, the larger issue involves all the labs who performed mixture interpretations “the old way” prior to the 2010 SWGDAM guideline changes. This includes about 99% of forensic laboratories in the United States. Those mixtures may not have been interpreted using the most accurate or conservative methods and they may have falsely implicated or included a person of interest.
I’ll let that sink in for a bit. How many thousands of cases out there might be impacted by this? The number is unknown, but is guaranteed to be high. In some of these cases, using appropriate statistical methods may yield very similar results, but in some case – especially those with multiple contributors and low levels of DNA – the results could be vastly different. That is what was found in Texas when laboratories began re-interpreting their mixtures. What might happen if other labs in other states begin taking a look at their old cases and reinterpreting the mixtures?
We can begin to have some idea of the impact by examining the Texas cases, as well as anecdotal results from various cases in labs around the country. While I, as a forensic DNA consultant, have had the opportunity to review a number of cases where re-interpretation of mixtures have yielded very different results, not everyone has the same access or information. As noted in Texas, results can change vastly when new interpretation methods are used on the same DNA data. It is not unusual to have a CPI change from 1 in several hundred thousand to 1 in 100. Even worse, results that once implicated the suspect can change from an inclusion to uninterpretable.
But Aren’t Forensic Laboratories Audited?
You may be asking yourself, how could this happen when laboratories are audited by outside agencies in order to attain accreditation? Would the auditors not flag laboratories who did not begin complying quickly with the 2010 SWGDAM guidelines? Would the audits not address the hundreds or thousands of cases involving mixtures which were incorrectly or inappropriately interpreted? The short answer is, no they would not. First, the audit is only as good as the auditor. And, frankly, the auditors look at only a tiny fraction of the cases that have been tested over the prior year or two. These cases may even have been selected by the laboratory under audit. There are many other areas that auditors are tasked with reviewing, so actual case file review is minimal. In regards to mixture issues, SWGDAM is tasked only with providing guidance to the forensic DNA community. Laboratories do not have to follow SWGDAM guidelines as they are not incorporated into the FBI’s Quality Assurance Standards for Forensic DNA Testing Laboratories, which all accredited forensic labs are audited against on a annual basis. If the lab chose not to update their mixture interpretation protocols in order to comply with SWGDAM guidelines, there was nothing that the auditors could do about it.
That began to change in July of 2015 with the issuance of the American Society of Crime Laboratory Directors- Laboratory Accreditation Board (ASCLD-LAB – one of the accrediting bodies in the forensic community at that time) Mixture Interpretation Position which stated that labs must have a written mixture interpretation procedure that is based on validation data and contains specific defined steps such that two individuals at the same lab would reach the same conclusion when interpreting a mixture profile. The procedure must also be tested on laboratory-created mixture of known contributors with varying number of contributors, varying proportions, and varying template quantities. This position statement, in combination with the temporary loss of accreditation and closure of the DC Consolidated Laboratory the prior April, ensured that ASCLD-LAB accredited laboratories would take a much closer look at their own mixture interpretation protocols in an effort to ensure that they can actually give an accurate answer to the question of “Is the person of interest a contributor to this mixture?”
Where Does This Leave Us?
So, where does this leave us? Fortunately, forensic DNA laboratories are recognizing that they do indeed need to follow SWGDAM’s mixture interpretation guidelines. It is rare for me to review a case that was processed in 2015 or later and find that the lab has not properly applied the CPI statistic to a mixed DNA profile. In addition, more and more labs are turning to probabilistic genotyping software to aid them in their interpretation of complex, low-level mixtures that are commonplace in today’s forensic DNA laboratories. While the emphasis on proper interpretation of mixtures is great news for current cases coming through the DNA lab’s door, this still leaves the unanswered question of “What do we do with all those cases that were improperly interpreted?”. Many in the field have a pat answer of “The SWGDAM guidelines state that they are not to be applied retroactively”. While it is true that the SWGDAM does indeed warn against a retroactive application of their guidelines, with all due respect to this highly beneficial organization, who is SWGDAM to say that the guidelines shouldn’t be applied retroactively? Shouldn’t that be a question for lawyers, judges, and those many individuals who have been tried and imprisoned based upon potentially faulty interpretation of mixture data?
If we now know that a method that was used in the past had the possibility of leading to less than conservative results and was potentially overly-inclusive – meaning there could be false positive inclusions and, therefore, the potential for falsely imprisoned individuals – shouldn’t laboratories around the country, not just in Texas, take a closer look at their older cases? I, for one, think so.
References:
NIST 2005 “MIX05 Interlaboratory Study” poster summarizing data can be found at: http:// www.cstl.nist.gov/strbase/interlab/MIX05.htm. Presentation by Dr. John Butler entitled “Mixture Interpretation: Lessons Learned from the MIX05 Interlaboratory Study” can be found at: http:/www.cstl.nist.gov/biotech/strbase/NISTpug.htm
SWGDAM Interpretation Guidelines for Autosomal STR Typing by Forensic DNA Testing ?Laboratories, 2010. http://www.fbi.gov/about-us/lab/biometric-analysis/codis/swgdam- ?interpretation-guidelines.
Dror IE and Hampikian G. Subjectivity and bias in forensic DNA mixture interpretation. ?Science and Justice 51(2011) 204-208.
Bieber F et al. Evaluation of forensic DNA mixture evidence: protocol for evaluation, interpretation, and statistical calculations using the combined probability of inclusion. BioMed Central Genetics (2016) 17:125
Curran, JM and Buckleton, J. Inclusion Probabilities and Dropout. J. Forensic Sci. September 2010, Vol. 55, No. 5; 1171-1173